Introduction
Congenital abnormalities account for 20-25% of perinatal deaths. Now, many genetic and other disorders can be diagnosed early in pregnancy.
Prenatal diagnosis uses various noninvasive and invasive techniques to determine the health of, the condition of, or any abnormality in an unborn fetus. These techniques are outlined below.
Noninvasive techniques
Ultrasound is a noninvasive procedure for imaging fetal anatomy. It is harmless to both the fetus and the mother. Ultrasound can evaluate gestational age, as well as identify twins; fetal position; placental location; fetal growth, development, and movement; and any structural birth defects. It also can assess amniotic fluid volume.
Many fetal organ systems and anatomical lesions, including some genitourinary, gastrointestinal, skeletal, and central nervous system abnormalities and congenital cardiopathies, can be visualized by ultrasound between 16-20 weeks' gestation. Using ultrasound, Romain et al1 carried out prenatal detection of congenital cataract in an unborn fetus with trisomy 21. Aslan et al2 carried out prenatal diagnosis of Neu-Laxova syndrome at 38 weeks' gestation. Ultrasound also is used to guide invasive sampling, such as amniocentesis, CVS, cordocentesis, and various fetal biopsies.
Fetal visualization - Fetal echocardiography
Fetal echocardiography can be performed at 15 weeks' gestation and beyond. When this technique is used with duplex or color flow Doppler, it can identify a number of major structural cardiac defects and rhythm disturbances.3 By fetal echocardiography, Saygili et al4 diagnosed absent pulmonary valve syndrome in a male infant prenatally at 22 weeks' gestation. Fetal echocardiography is recommended in cases where cardiac defects are suspected, including the following:
MRI is a fetal imaging technique that uses powerful magnets and radio waves to construct images of the body, but, because of fetal movements, its application has been limited. Glenn et al reported a prenatal diagnosis of polymicrogyria using MRI as an imaging technique.5
Fetal visualization - Radiography
The fetal skeleton can be visualized by radiography from 10 weeks' gestation onward. This technique is used for the diagnosis of inherited skeletal dysplasias, particularly osteochondrodysplasia, in the second and third trimesters. Aslan et al2 carried out prenatal diagnosis of thanatophoric dysplasia at 19 weeks' gestation in an 18-year-old woman. Because of the dangers of radiography to the fetus, this technique rarely is used.
Screening for neural tube defects
Screening for NTDs is recommended if the following are present:
The developing fetus has 2 major blood proteins, albumin and alpha-fetoprotein (AFP), while adults have only albumin in their blood. The MSAFP level can be used to determine the AFP levels from the fetus. AFP is produced by the yolk sac and later by the liver; it enters the amniotic fluid and then the maternal serum via fetal urine.
In the condition of an open NTD (eg, anencephaly, spina bifida) and abdominal wall defects in the fetus, AFP diffuses rapidly from exposed fetal tissues into amniotic fluid, and the MSAFP level rises. However, the MSAFP levels also increase with gestational age, gestational diabetes, twins, pregnancies complicated by bleeding, and in association with intrauterine growth retardation.
The MSAFP test has the greatest sensitivity between 16-18 weeks' gestation, but it also can be performed between 15-22 weeks' gestation. A combination of the MSAFP test and ultrasonography detects almost all cases of anencephaly and most cases of spina bifida. Also, a NTD can be distinguished from other fetal defects, such as abdominal wall defects, by the use of an acetylcholinesterase test carried out on amniotic fluid obtained by amniocentesis. If the level of acetylcholinesterase rises along with AFAFP, it is suspected as a condition of a NTD.6
Screening for fetal Down syndrome - Measuring maternal serum alpha-fetoprotein
In cases where a low level of MSAFP is reported, it indicates the condition of Down syndrome or other chromosomal aneuploidy and failing pregnancies.7,8
Screening for fetal Down syndrome - Measuring maternal unconjugated estriol
The amount of estriol in maternal serum depends upon viable fetus, a properly functioning placenta, and on maternal well-being. Fetal adrenal glands produce dehydroepiandrosterone (DHEA) that gets metabolized to estriol in the placenta. Estriol crosses to the maternal circulation and is excreted either by maternal kidney in urine or by maternal liver in the bile. In the third trimester, the level of estriol gives an indication for the well-being of the fetus. A low level of estriol is an indication of Down syndrome and adrenal hyperplasia with anencephaly.9,10 If the estriol level drops to a great level, then it indicates risk to fetus.
Screening for fetal Down syndrome - Measuring maternal serum beta-human chorionic gonadotropin
Following conception and implantation of the developing embryo into the uterus, the trophoblasts produce enough beta-HCG, which is an indication for pregnancy. In the middle to late second trimester, the level of beta-HCG also can be used in conjunction with the MSAFP level to screen for chromosomal abnormalities. An increased beta-HCG level coupled with a decreased MSAFP level suggests Down syndrome.11,8 The beta-HCG level also can be quantified in serum from maternal blood, and, if its amount is found to be lower than expected, it indicates abortion or ectopic pregnancy. If the level of HCG is estimated to be considerably high, then it indicates the possibility of trophoblastic diseases. The elevated level of HCG, along with absence of the fetus on ultrasonography, indicates a hydatidiform mole.
Screening for fetal Down syndrome - Measuring maternal inhibin-A levels
The hormone inhibin is secreted by the placenta and the corpus luteum. Inhibin-A can be measured in maternal serum. An increased level of inhibin-A is linked with an increased risk for trisomy 21. A high inhibin-A level may also be associated with a risk for preterm delivery.
Screening for fetal Down syndrome - Cell-free fetal nucleic acids from the placenta
Cell-free fetal DNA and RNA can be extracted from maternal blood around 7 weeks’ gestation, which can be used to screen for Down syndrome. Sex determination for families with inherited sex-linked diseases, diagnosis of certain single gene disorders, and blood Rhesus factor status (in the case of Rhesus D-negative mothers) can also be performed using cell-free fetal nucleic acids from the placenta.
Screening for fetal Down syndrome - Separation of fetal cells from the mother's blood
Fetal blood cells make access to maternal circulation through the placental villi. These cells can be collected safely from approximately 18 weeks' gestation onward, although by successful procedures, these cells can be collected at 12 weeks' gestation.12 The fetal cells can be sorted out and analyzed by different techniques.
Fluorescent in situ hybridization (FISH) is one technique that can be used to diagnose aneuploid conditions, such as trisomies and monosomy X. In the condition of fetal infection with such viruses as rubella, cytomegalovirus, and toxoplasmosis, the viral immunoglobulin M (IgM) or DNA also can be identified in fetal blood.13,14
Fetal blood cells can be analyzed for the diagnosis of genetic disorders using molecular genetic techniques by isolating DNA and amplifying it by polymerase chain reaction (PCR).
Fetal cells separated from a mother's blood have been successfully used in the diagnosis of cystic fibrosis, sickle cell anemia, and thalassemia in a fetus.
Embryoscopy is performed in the first trimester of pregnancy (up to 12 weeks’ gestation).15 In this technique, a rigid endoscope is inserted via the cervix in the space between the amnion and the chorion, under sterile conditions and ultrasound guidance, to visualize the embryo for the diagnosis of structural malformations.
Fetal visualization - Fetoscopy
Fetoscopy is performed during the second trimester (after 16 weeks’ gestation). In this technique, a fine-caliber endoscope is inserted into the amniotic cavity through a small maternal abdominal incision, under sterile conditions and ultrasound guidance, for the visualization of the embryo to detect the presence of subtle structural abnormalities. It also is used for fetal blood and tissue sampling. Fetoscopy is associated with a 3-5% risk of miscarriage; therefore, it is superseded by detailed ultrasound scanning.
Fetal tissue sampling - Amniocentesis
Amniocentesis is an invasive, well-established, safe, reliable, and accurate procedure performed between 14-20 weeks of pregnancy. Amniocentesis is advised for pregnant women at 35 years or older for detection of chromosomal abnormalities in the fetus.
It is performed under ultrasound guidance. A 22-gauge needle is passed through the mother's lower abdomen into the amniotic cavity inside the uterus, and 10-20 mL of amniotic fluid that contains cells from amnion, fetal skin, fetal lungs, and urinary tract epithelium are collected. These cells are grown in culture for chromosomal, biochemical, and molecular biologic analyses. Supernatant amniotic fluid is used for the measurement of substances, such as AFAFP, hormones, and enzymes.
The results of cytogenetic and biochemical studies on amniotic cell cultures are more than 90% accurate. In the third trimester of pregnancy, the amniotic fluid can be analyzed for determination of fetal lung maturity. Risks with amniocentesis are rare but include 0.5-1.0% fetal loss and maternal Rh sensitization.
Fetal tissue sampling - Chorionic villus sampling
CVS is performed very early in gestation between 9-12 weeks, ideally at 10 weeks' gestation. A catheter is passed through the cervix or through the abdominal wall into the uterus under ultrasound guidance, and a sample of chorionic villi surrounding the sac is obtained. The villi are dissected from the decidual tissue, and chromosome analysis is carried out on these cells to determine the karyotype of the fetus (see image below).
DNA can be extracted from these cells for molecular analysis. DNA analysis of CVS specimens is helpful for early diagnosis of hemoglobinopathies.16 In addition, tissue culture can be initiated on these cells for further studies.
The major advantage of CVS over amniocentesis is getting quick results and its use in early pregnancy. Abnormalities can be identified at an early stage, and more acceptable decisions about termination of the pregnancy can be taken. Abortion is also much safer at this early stage. A disadvantage of CVS as compared to amniocentesis is a 2-3% risk of causing miscarriage, and, rarely, CVS can result with limb defects in the fetus.17 Maternal sensitization is possible. A higher rate of maternal cell contamination and confined placental mosaicism with CVS may result in diagnostic ambiguity, leading to the need for additional invasive diagnostic tests.18
Fetal tissue sampling - Percutaneous umbilical blood sampling
PUBS is also known as cordocentesis.14 It is a method for fetal blood sampling and is performed after 16 weeks' gestation. A needle is inserted into the umbilical cord under ultrasound guidance, and fetal blood is collected from the umbilical vein for chromosome analysis and genetic diagnosis. An advantage of PUBS is the rapid rate at which lymphocytes grow, allowing prompt genetic diagnosis.
This technique is also useful for evaluating fetal metabolism and hematologic abnormalities.
Fetal tissue sampling - Percutaneous skin biopsy
To prenatally diagnose a number of serious skin disorders, such as anhidrotic ectodermal dysplasia, epidermolysis bullosa letalis, epidermolysis bullosa dystrophica, hypohidrotic ectodermal dysplasia, oculocutaneous albinism, and genetic forms of ichthyosis, percutaneous fetal skin biopsies are taken under ultrasonic guidance between 17-20 weeks' gestation.
Fetal tissue sampling - Other organ biopsies, including liver and muscle biopsy
Fetal liver biopsy is needed to diagnose an inborn error of metabolism, such as ornithine transcarbamylase deficiency19 , glucose-6-phosphatase deficiency20 , glycogen storage disease type IA, nonketotic hyperglycemia21 , and carbamoyl-phosphate synthetase deficiency.22 Fetal liver biopsy also is best performed between 17-20 weeks' gestation under ultrasound guidance.
Fetal muscle biopsy is carried out under ultrasound guidance at about 18 weeks' gestation to analyze the muscle fibers histochemically for prenatal diagnosis of Becker-Duchenne muscular dystrophy.23
Fetal tissue sampling - Preimplantation biopsy of blastocysts obtained by in vitro fertilization
Techniques are being developed to test cells obtained from biopsy of early cleavage stages or blastocysts of pregnancies conceived through in vitro fertilization.24 These techniques will be helpful for selective transfer and implantation of those pregnancies into the uterus that are not affected by a specific genetic disorder. This approach will be more acceptable to those couples who oppose abortions.
Chromosomal aberrations, such as deletions, duplications, translocations, and inversions diagnosed in affected parents or siblings, can be detected prenatally in a fetus by chromosomal analysis (see image below).
This analysis can be undertaken on fetal cells obtained through such techniques as amniocentesis and CVS.
Fluorescent in situ hybridization
FISH uses different fluorescent-labeled probes, which are single-stranded DNA conjugated with fluorescent dyes and are specific to regions of individual chromosomes. These probes hybridize with complementary target DNA sequences25 in the genome and can detect chromosomal abnormalities, such as trisomies26 , monosomies, and duplications.
Three types of DNA probes are used in FISH analysis. Whole chromosome probes are specific to a whole chromosome or a chromosome segment and are applied to metaphase spread for the identification of translocations or aneuploidy. Repetitive probes, such as alpha satellite sequences located in the centromeric regions of human chromosomes, are used in the identification of marker chromosomes and aneuploidy. Unique sequence probes are single clones or a series of overlapping clones corresponding to a specific gene or a confined region of a chromosome that do not contain major repetitive sequences and are used for the identification of specific translocation events in cancer27 and for the detection of submicroscopic deletions.28
In 4% of retinoblastoma cases, deletion of chromosome band 13q14 has been reported.29 Prenatal diagnosis of retinoblastoma cases with deletion of this band on chromosome 13 is feasible using fluorescent-labeled probes for this region. Hybridization of fluorescent DNA probes to interphase nuclei is under investigation as a screening method for aneuploidy.
Microarray comparative genomic hybridization
Recently, array-CGH (microarray comparative genomic hybridization) is considered to be useful in detecting genomic imbalance in the fetus (duplications/deletions).
Molecular genetic techniques are being used for prenatal diagnosis.30 These techniques are based upon the fact that DNA complement is generally identical in every cell of the body; therefore, any hereditary defect diagnosed at the DNA level will be present in nucleated cells from that individual. For molecular analysis, DNA is extracted from amniocytes, chorionic villi, or fetal blood cells. Then, it is amplified by PCR and is used for the diagnosis of genetic mutations or deletions within a gene that causes a specific genetic disease. The following molecular biologic techniques can be used for prenatal diagnosis of different diseases.
Linkage analysis by microsatellite markers
Microsatellites are short tandem repeats of 2-6 base pairs that are highly polymorphic and are distributed throughout the genome. This form of polymorphism is inherited in a mendelian codominant manner. For linkage analysis, primers for regions flanking the repeat sequences are designed and used to amplify these microsatellites by PCR, initially for candidate gene regions and on their exclusion for whole genome analysis.
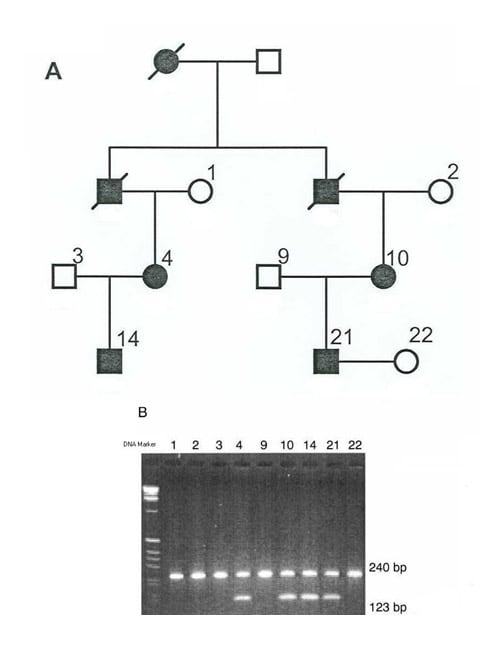
On gel electrophoresis, the genotype of different individuals in the family indicating 2 alleles for each microsatellite marker is established, and haplotypes are constructed with the analyzed markers. Cosegregation of a particular allele of any of these analyzed markers with the disease phenotype, in all the affected but in none of the unaffected individuals, indicates the probability of linkage with that marker at that particular locus, which is confirmed statistically by calculating the lod scores. A lod score value of greater than 3 indicates linkage of that particular marker with the disease locus in that family. In informative families affected with a disease, linkage can be confirmed by lod score and haplotype analysis. Segregation of a particular allele linked with disease phenotype also can be tested in the fetus by haplotype analysis (see image below).
Carter et al31 identified an intragenic polymorphic marker linked with human CP49 gene (that codes for intermediate filament protein in lens fiber cells) on chromosome 3 at band 3q21-22 for the genetic linkage analysis of autosomal dominant congenital cataract. Toudjarska et al32 demonstrated molecular diagnosis of Marfan syndrome by linkage analysis.
Restriction fragment length polymorphism
In the human genome, variations are common and reportedly occur approximately once every 200 base pairs. These single base pair differences in DNA nucleotide sequences are inherited in a mendelian codominant manner. Restriction endonucleases are the enzymes that recognize and cut DNA within a specific base sequence recognition site. If a difference occurs in the DNA sequence within the recognition sequence of a restriction enzyme, it results in fragments of different size by that restriction enzyme. This difference is recognized by the altered mobility of the restriction fragments on gel electrophoresis, which is known as RFLP (see image below). This technique is used to detect deletions within the gene and DNA polymorphisms and to identify mutant genes and mutations at hot spots.
Churchill et al33 performed prenatal diagnosis in a familial case of aniridia by extracting DNA from cultured fibroblasts obtained through amniocentesis, RFLP with restriction enzyme Ava1, and electrophoresis by single-strand confirmation polymorphism to screen the PAX6 gene.
Single nucleotide polymorphisms
SNPs are single base differences in the genome of an individual, which occur about every 1000 bases. Each SNP has 2 alleles; they can be used for linkage analysis to carry out fine mapping of regions on the chromosomes and to study mutations in the genes. The advantages of SNPs are their abundant numbers, and they can be typed by oligonucleotide hybridization assay, without gel electrophoresis. Two methods are available for oligonucleotide hybridization assay, DNA chip and DASH.
DNA chip
A DNA chip is a wafer of silicon, usually 2 cm3 or less in area, and carries many different oligonucleotides (short single-stranded DNA molecule less than 50 nucleotides in length synthesized artificially in a test tube) in a high-density array. The DNA to be analyzed is labeled with a fluorescent marker and is pipetted onto the surface of the chip. Hybridization of labeled DNA is detected by examining the chip with a fluorescent microscope. The position where the hybridization signal is emitted indicates which oligonucleotide has hybridized with the test DNA. If there is a single mismatch at a single position within the oligonucleotide, that mismatch does not form a base pair, and hybridization does not occur. In this way, oligonucleotide hybridization discriminates between the 2 alleles of a SNP.
Dynamic allele-specific hybridization
In this technique, hybridization takes place in solution, in 1 of the 96 well microtiter tray. Hybridization is detected by a fluorescent marker that binds only to double-stranded DNA and emits a signal on hybridization. Initially, hybridization is carried out under conditions that allow mismatched hybrids to form, and, at this stage, oligonucleotides and the test DNA hybridize regardless of which SNP allele is contained by the DNA. By raising the temperature, the mismatched hybrids, which are less stable as compared to complete hybrids, break down. Detecting which allele is present in the test DNA can be determined from the temperature at which the hybridization-dependent fluorescent signal disappears.
Currently, SNPs are used for the molecular genetic analysis of many eye disorders, such as congenital cataract, myopia, Marfan syndrome, and glaucoma.
Congenital abnormalities account for 20-25% of perinatal deaths. Now, many genetic and other disorders can be diagnosed early in pregnancy.
Prenatal diagnosis uses various noninvasive and invasive techniques to determine the health of, the condition of, or any abnormality in an unborn fetus. These techniques are outlined below.
Noninvasive techniques
- Fetal visualization
- Ultrasound
- Fetal echocardiography
- Magnetic resonance imaging (MRI)
- Radiography
- Screening for neural tube defects (NTDs) - Measuring maternal serum alpha-fetoprotein (MSAFP)
- Screening for fetal Down syndrome
- Measuring MSAFP
- Measuring maternal unconjugated estriol
- Measuring maternal serum beta-human chorionic gonadotropin (HCG)
- Separation of fetal cells from the mother's blood
- Fetal visualization
- Embryoscopy
- Fetoscopy
- Fetal tissue sampling
- Amniocentesis
- Chorionic villus sampling (CVS)
- Percutaneous umbilical blood sampling (PUBS)
- Percutaneous skin biopsy
- Other organ biopsies, including muscle and liver biopsy
- Preimplantation biopsy of blastocysts obtained by in vitro fertilization
- Cytogenetic investigations
- Detection of chromosomal aberrations
- Fluorescent in situ hybridization
- Molecular genetic techniques
- Linkage analysis using microsatellite markers
- Restriction fragment length polymorphisms (RFLPs)
- Single nucleotide polymorphisms (SNPs)
- DNA chip
- Dynamic allele-specific hybridization (DASH)
Noninvasive Techniques
Fetal visualization - UltrasoundUltrasound is a noninvasive procedure for imaging fetal anatomy. It is harmless to both the fetus and the mother. Ultrasound can evaluate gestational age, as well as identify twins; fetal position; placental location; fetal growth, development, and movement; and any structural birth defects. It also can assess amniotic fluid volume.
Many fetal organ systems and anatomical lesions, including some genitourinary, gastrointestinal, skeletal, and central nervous system abnormalities and congenital cardiopathies, can be visualized by ultrasound between 16-20 weeks' gestation. Using ultrasound, Romain et al1 carried out prenatal detection of congenital cataract in an unborn fetus with trisomy 21. Aslan et al2 carried out prenatal diagnosis of Neu-Laxova syndrome at 38 weeks' gestation. Ultrasound also is used to guide invasive sampling, such as amniocentesis, CVS, cordocentesis, and various fetal biopsies.
Fetal visualization - Fetal echocardiography
Fetal echocardiography can be performed at 15 weeks' gestation and beyond. When this technique is used with duplex or color flow Doppler, it can identify a number of major structural cardiac defects and rhythm disturbances.3 By fetal echocardiography, Saygili et al4 diagnosed absent pulmonary valve syndrome in a male infant prenatally at 22 weeks' gestation. Fetal echocardiography is recommended in cases where cardiac defects are suspected, including the following:
- Identification of an extracardiac malformation on routine ultrasound
- Abnormality of another major organ system
- Suspected genetic disease or fetal chromosome abnormality associated with heart defects
- Exposure to potentially teratogenic agents
- Family history of congenital heart defects, particularly in a parent or sibling
- Maternal diseases, such as diabetes or phenylketonuria associated with fetal structural heart defects, in particular heart blocks, such as lupus or other immune disorders
- Alcohol or drug consumption by mother during pregnancy
- Maternal rubella infection during pregnancy
MRI is a fetal imaging technique that uses powerful magnets and radio waves to construct images of the body, but, because of fetal movements, its application has been limited. Glenn et al reported a prenatal diagnosis of polymicrogyria using MRI as an imaging technique.5
Fetal visualization - Radiography
The fetal skeleton can be visualized by radiography from 10 weeks' gestation onward. This technique is used for the diagnosis of inherited skeletal dysplasias, particularly osteochondrodysplasia, in the second and third trimesters. Aslan et al2 carried out prenatal diagnosis of thanatophoric dysplasia at 19 weeks' gestation in an 18-year-old woman. Because of the dangers of radiography to the fetus, this technique rarely is used.
Screening for neural tube defects
Screening for NTDs is recommended if the following are present:
- Ultrasound findings indicate NTDs.
- A child with NTDs is already in the family.
- A family history of NTDs exists, especially a mother with NTDs.
- The mother has type 1 diabetes mellitus during pregnancy.
- Maternal exposure to drugs, such as valproic acid, is associated with NTDs.
- Elevated level of MSAFP is present.
The developing fetus has 2 major blood proteins, albumin and alpha-fetoprotein (AFP), while adults have only albumin in their blood. The MSAFP level can be used to determine the AFP levels from the fetus. AFP is produced by the yolk sac and later by the liver; it enters the amniotic fluid and then the maternal serum via fetal urine.
In the condition of an open NTD (eg, anencephaly, spina bifida) and abdominal wall defects in the fetus, AFP diffuses rapidly from exposed fetal tissues into amniotic fluid, and the MSAFP level rises. However, the MSAFP levels also increase with gestational age, gestational diabetes, twins, pregnancies complicated by bleeding, and in association with intrauterine growth retardation.
The MSAFP test has the greatest sensitivity between 16-18 weeks' gestation, but it also can be performed between 15-22 weeks' gestation. A combination of the MSAFP test and ultrasonography detects almost all cases of anencephaly and most cases of spina bifida. Also, a NTD can be distinguished from other fetal defects, such as abdominal wall defects, by the use of an acetylcholinesterase test carried out on amniotic fluid obtained by amniocentesis. If the level of acetylcholinesterase rises along with AFAFP, it is suspected as a condition of a NTD.6
Screening for fetal Down syndrome - Measuring maternal serum alpha-fetoprotein
In cases where a low level of MSAFP is reported, it indicates the condition of Down syndrome or other chromosomal aneuploidy and failing pregnancies.7,8
Screening for fetal Down syndrome - Measuring maternal unconjugated estriol
The amount of estriol in maternal serum depends upon viable fetus, a properly functioning placenta, and on maternal well-being. Fetal adrenal glands produce dehydroepiandrosterone (DHEA) that gets metabolized to estriol in the placenta. Estriol crosses to the maternal circulation and is excreted either by maternal kidney in urine or by maternal liver in the bile. In the third trimester, the level of estriol gives an indication for the well-being of the fetus. A low level of estriol is an indication of Down syndrome and adrenal hyperplasia with anencephaly.9,10 If the estriol level drops to a great level, then it indicates risk to fetus.
Screening for fetal Down syndrome - Measuring maternal serum beta-human chorionic gonadotropin
Following conception and implantation of the developing embryo into the uterus, the trophoblasts produce enough beta-HCG, which is an indication for pregnancy. In the middle to late second trimester, the level of beta-HCG also can be used in conjunction with the MSAFP level to screen for chromosomal abnormalities. An increased beta-HCG level coupled with a decreased MSAFP level suggests Down syndrome.11,8 The beta-HCG level also can be quantified in serum from maternal blood, and, if its amount is found to be lower than expected, it indicates abortion or ectopic pregnancy. If the level of HCG is estimated to be considerably high, then it indicates the possibility of trophoblastic diseases. The elevated level of HCG, along with absence of the fetus on ultrasonography, indicates a hydatidiform mole.
Screening for fetal Down syndrome - Measuring maternal inhibin-A levels
The hormone inhibin is secreted by the placenta and the corpus luteum. Inhibin-A can be measured in maternal serum. An increased level of inhibin-A is linked with an increased risk for trisomy 21. A high inhibin-A level may also be associated with a risk for preterm delivery.
Screening for fetal Down syndrome - Cell-free fetal nucleic acids from the placenta
Cell-free fetal DNA and RNA can be extracted from maternal blood around 7 weeks’ gestation, which can be used to screen for Down syndrome. Sex determination for families with inherited sex-linked diseases, diagnosis of certain single gene disorders, and blood Rhesus factor status (in the case of Rhesus D-negative mothers) can also be performed using cell-free fetal nucleic acids from the placenta.
Screening for fetal Down syndrome - Separation of fetal cells from the mother's blood
Fetal blood cells make access to maternal circulation through the placental villi. These cells can be collected safely from approximately 18 weeks' gestation onward, although by successful procedures, these cells can be collected at 12 weeks' gestation.12 The fetal cells can be sorted out and analyzed by different techniques.
Fluorescent in situ hybridization (FISH) is one technique that can be used to diagnose aneuploid conditions, such as trisomies and monosomy X. In the condition of fetal infection with such viruses as rubella, cytomegalovirus, and toxoplasmosis, the viral immunoglobulin M (IgM) or DNA also can be identified in fetal blood.13,14
Fetal blood cells can be analyzed for the diagnosis of genetic disorders using molecular genetic techniques by isolating DNA and amplifying it by polymerase chain reaction (PCR).
Fetal cells separated from a mother's blood have been successfully used in the diagnosis of cystic fibrosis, sickle cell anemia, and thalassemia in a fetus.
Invasive Techniques
Fetal visualization -EmbryoscopyEmbryoscopy is performed in the first trimester of pregnancy (up to 12 weeks’ gestation).15 In this technique, a rigid endoscope is inserted via the cervix in the space between the amnion and the chorion, under sterile conditions and ultrasound guidance, to visualize the embryo for the diagnosis of structural malformations.
Fetal visualization - Fetoscopy
Fetoscopy is performed during the second trimester (after 16 weeks’ gestation). In this technique, a fine-caliber endoscope is inserted into the amniotic cavity through a small maternal abdominal incision, under sterile conditions and ultrasound guidance, for the visualization of the embryo to detect the presence of subtle structural abnormalities. It also is used for fetal blood and tissue sampling. Fetoscopy is associated with a 3-5% risk of miscarriage; therefore, it is superseded by detailed ultrasound scanning.
Fetal tissue sampling - Amniocentesis
Amniocentesis is an invasive, well-established, safe, reliable, and accurate procedure performed between 14-20 weeks of pregnancy. Amniocentesis is advised for pregnant women at 35 years or older for detection of chromosomal abnormalities in the fetus.
It is performed under ultrasound guidance. A 22-gauge needle is passed through the mother's lower abdomen into the amniotic cavity inside the uterus, and 10-20 mL of amniotic fluid that contains cells from amnion, fetal skin, fetal lungs, and urinary tract epithelium are collected. These cells are grown in culture for chromosomal, biochemical, and molecular biologic analyses. Supernatant amniotic fluid is used for the measurement of substances, such as AFAFP, hormones, and enzymes.
The results of cytogenetic and biochemical studies on amniotic cell cultures are more than 90% accurate. In the third trimester of pregnancy, the amniotic fluid can be analyzed for determination of fetal lung maturity. Risks with amniocentesis are rare but include 0.5-1.0% fetal loss and maternal Rh sensitization.
Fetal tissue sampling - Chorionic villus sampling
CVS is performed very early in gestation between 9-12 weeks, ideally at 10 weeks' gestation. A catheter is passed through the cervix or through the abdominal wall into the uterus under ultrasound guidance, and a sample of chorionic villi surrounding the sac is obtained. The villi are dissected from the decidual tissue, and chromosome analysis is carried out on these cells to determine the karyotype of the fetus (see image below).
DNA can be extracted from these cells for molecular analysis. DNA analysis of CVS specimens is helpful for early diagnosis of hemoglobinopathies.16 In addition, tissue culture can be initiated on these cells for further studies.
The major advantage of CVS over amniocentesis is getting quick results and its use in early pregnancy. Abnormalities can be identified at an early stage, and more acceptable decisions about termination of the pregnancy can be taken. Abortion is also much safer at this early stage. A disadvantage of CVS as compared to amniocentesis is a 2-3% risk of causing miscarriage, and, rarely, CVS can result with limb defects in the fetus.17 Maternal sensitization is possible. A higher rate of maternal cell contamination and confined placental mosaicism with CVS may result in diagnostic ambiguity, leading to the need for additional invasive diagnostic tests.18
Fetal tissue sampling - Percutaneous umbilical blood sampling
PUBS is also known as cordocentesis.14 It is a method for fetal blood sampling and is performed after 16 weeks' gestation. A needle is inserted into the umbilical cord under ultrasound guidance, and fetal blood is collected from the umbilical vein for chromosome analysis and genetic diagnosis. An advantage of PUBS is the rapid rate at which lymphocytes grow, allowing prompt genetic diagnosis.
This technique is also useful for evaluating fetal metabolism and hematologic abnormalities.
Fetal tissue sampling - Percutaneous skin biopsy
To prenatally diagnose a number of serious skin disorders, such as anhidrotic ectodermal dysplasia, epidermolysis bullosa letalis, epidermolysis bullosa dystrophica, hypohidrotic ectodermal dysplasia, oculocutaneous albinism, and genetic forms of ichthyosis, percutaneous fetal skin biopsies are taken under ultrasonic guidance between 17-20 weeks' gestation.
Fetal tissue sampling - Other organ biopsies, including liver and muscle biopsy
Fetal liver biopsy is needed to diagnose an inborn error of metabolism, such as ornithine transcarbamylase deficiency19 , glucose-6-phosphatase deficiency20 , glycogen storage disease type IA, nonketotic hyperglycemia21 , and carbamoyl-phosphate synthetase deficiency.22 Fetal liver biopsy also is best performed between 17-20 weeks' gestation under ultrasound guidance.
Fetal muscle biopsy is carried out under ultrasound guidance at about 18 weeks' gestation to analyze the muscle fibers histochemically for prenatal diagnosis of Becker-Duchenne muscular dystrophy.23
Fetal tissue sampling - Preimplantation biopsy of blastocysts obtained by in vitro fertilization
Techniques are being developed to test cells obtained from biopsy of early cleavage stages or blastocysts of pregnancies conceived through in vitro fertilization.24 These techniques will be helpful for selective transfer and implantation of those pregnancies into the uterus that are not affected by a specific genetic disorder. This approach will be more acceptable to those couples who oppose abortions.
Cytogenetic Investigations
Detection of chromosomal aberrationsChromosomal aberrations, such as deletions, duplications, translocations, and inversions diagnosed in affected parents or siblings, can be detected prenatally in a fetus by chromosomal analysis (see image below).
This analysis can be undertaken on fetal cells obtained through such techniques as amniocentesis and CVS.
Fluorescent in situ hybridization
FISH uses different fluorescent-labeled probes, which are single-stranded DNA conjugated with fluorescent dyes and are specific to regions of individual chromosomes. These probes hybridize with complementary target DNA sequences25 in the genome and can detect chromosomal abnormalities, such as trisomies26 , monosomies, and duplications.
Three types of DNA probes are used in FISH analysis. Whole chromosome probes are specific to a whole chromosome or a chromosome segment and are applied to metaphase spread for the identification of translocations or aneuploidy. Repetitive probes, such as alpha satellite sequences located in the centromeric regions of human chromosomes, are used in the identification of marker chromosomes and aneuploidy. Unique sequence probes are single clones or a series of overlapping clones corresponding to a specific gene or a confined region of a chromosome that do not contain major repetitive sequences and are used for the identification of specific translocation events in cancer27 and for the detection of submicroscopic deletions.28
In 4% of retinoblastoma cases, deletion of chromosome band 13q14 has been reported.29 Prenatal diagnosis of retinoblastoma cases with deletion of this band on chromosome 13 is feasible using fluorescent-labeled probes for this region. Hybridization of fluorescent DNA probes to interphase nuclei is under investigation as a screening method for aneuploidy.
Microarray comparative genomic hybridization
Recently, array-CGH (microarray comparative genomic hybridization) is considered to be useful in detecting genomic imbalance in the fetus (duplications/deletions).
Molecular Genetic Techniques
OverviewMolecular genetic techniques are being used for prenatal diagnosis.30 These techniques are based upon the fact that DNA complement is generally identical in every cell of the body; therefore, any hereditary defect diagnosed at the DNA level will be present in nucleated cells from that individual. For molecular analysis, DNA is extracted from amniocytes, chorionic villi, or fetal blood cells. Then, it is amplified by PCR and is used for the diagnosis of genetic mutations or deletions within a gene that causes a specific genetic disease. The following molecular biologic techniques can be used for prenatal diagnosis of different diseases.
Linkage analysis by microsatellite markers
Microsatellites are short tandem repeats of 2-6 base pairs that are highly polymorphic and are distributed throughout the genome. This form of polymorphism is inherited in a mendelian codominant manner. For linkage analysis, primers for regions flanking the repeat sequences are designed and used to amplify these microsatellites by PCR, initially for candidate gene regions and on their exclusion for whole genome analysis.
On gel electrophoresis, the genotype of different individuals in the family indicating 2 alleles for each microsatellite marker is established, and haplotypes are constructed with the analyzed markers. Cosegregation of a particular allele of any of these analyzed markers with the disease phenotype, in all the affected but in none of the unaffected individuals, indicates the probability of linkage with that marker at that particular locus, which is confirmed statistically by calculating the lod scores. A lod score value of greater than 3 indicates linkage of that particular marker with the disease locus in that family. In informative families affected with a disease, linkage can be confirmed by lod score and haplotype analysis. Segregation of a particular allele linked with disease phenotype also can be tested in the fetus by haplotype analysis (see image below).
Carter et al31 identified an intragenic polymorphic marker linked with human CP49 gene (that codes for intermediate filament protein in lens fiber cells) on chromosome 3 at band 3q21-22 for the genetic linkage analysis of autosomal dominant congenital cataract. Toudjarska et al32 demonstrated molecular diagnosis of Marfan syndrome by linkage analysis.
Restriction fragment length polymorphism
In the human genome, variations are common and reportedly occur approximately once every 200 base pairs. These single base pair differences in DNA nucleotide sequences are inherited in a mendelian codominant manner. Restriction endonucleases are the enzymes that recognize and cut DNA within a specific base sequence recognition site. If a difference occurs in the DNA sequence within the recognition sequence of a restriction enzyme, it results in fragments of different size by that restriction enzyme. This difference is recognized by the altered mobility of the restriction fragments on gel electrophoresis, which is known as RFLP (see image below). This technique is used to detect deletions within the gene and DNA polymorphisms and to identify mutant genes and mutations at hot spots.
Churchill et al33 performed prenatal diagnosis in a familial case of aniridia by extracting DNA from cultured fibroblasts obtained through amniocentesis, RFLP with restriction enzyme Ava1, and electrophoresis by single-strand confirmation polymorphism to screen the PAX6 gene.
Single nucleotide polymorphisms
SNPs are single base differences in the genome of an individual, which occur about every 1000 bases. Each SNP has 2 alleles; they can be used for linkage analysis to carry out fine mapping of regions on the chromosomes and to study mutations in the genes. The advantages of SNPs are their abundant numbers, and they can be typed by oligonucleotide hybridization assay, without gel electrophoresis. Two methods are available for oligonucleotide hybridization assay, DNA chip and DASH.
DNA chip
A DNA chip is a wafer of silicon, usually 2 cm3 or less in area, and carries many different oligonucleotides (short single-stranded DNA molecule less than 50 nucleotides in length synthesized artificially in a test tube) in a high-density array. The DNA to be analyzed is labeled with a fluorescent marker and is pipetted onto the surface of the chip. Hybridization of labeled DNA is detected by examining the chip with a fluorescent microscope. The position where the hybridization signal is emitted indicates which oligonucleotide has hybridized with the test DNA. If there is a single mismatch at a single position within the oligonucleotide, that mismatch does not form a base pair, and hybridization does not occur. In this way, oligonucleotide hybridization discriminates between the 2 alleles of a SNP.
Dynamic allele-specific hybridization
In this technique, hybridization takes place in solution, in 1 of the 96 well microtiter tray. Hybridization is detected by a fluorescent marker that binds only to double-stranded DNA and emits a signal on hybridization. Initially, hybridization is carried out under conditions that allow mismatched hybrids to form, and, at this stage, oligonucleotides and the test DNA hybridize regardless of which SNP allele is contained by the DNA. By raising the temperature, the mismatched hybrids, which are less stable as compared to complete hybrids, break down. Detecting which allele is present in the test DNA can be determined from the temperature at which the hybridization-dependent fluorescent signal disappears.
Currently, SNPs are used for the molecular genetic analysis of many eye disorders, such as congenital cataract, myopia, Marfan syndrome, and glaucoma.
Prerequisites of Prenatal Diagnosis
Prenatal diagnosis is recommended in the following cases:- The pregnant woman is 35 years or older at the time of delivery.
- She or her parents have had a previous child with a chromosomal abnormality.
- She has a history of recurrent abortions, or her husband's previous wife experienced several miscarriages.
- A history of parental consanguinity is present.
- The couple is known to be carriers of a chromosomal translocation.
- The pregnant woman is affected with type 1 diabetes mellitus, epilepsy, or myotonic dystrophy.
- She is exposed to viral infections, such as rubella or cytomegalovirus.
- The mother is exposed to excessive medication or to environmental hazards.
- In her or her spouse's family, a history of Down syndrome or some other chromosomal abnormality is present.
- A history of single gene disorder is present in her or her spouse's family.
- Her male relatives have Duchenne muscular dystrophy or severe hemophilia.
- She is suspected of having some other harmful gene on her X chromosomes.
- The fetus is diagnosed in utero to have some hereditary error of metabolism.
- The fetus is detected to be at increased risk for a NTD.
Benefits of Prenatal Diagnosis
The benefits of prenatal diagnosis are as follows:- Prenatal diagnosis determines the outcome of pregnancy.
- It is helpful for couples to decide whether to continue the pregnancy.
- It indicates possible complications that can arise at birth process.
- Prenatal diagnosis is helpful for the management of remaining weeks of pregnancy.
- It prepares the couple for the birth of a child with an abnormality.
- Prenatal diagnosis can be helpful for the improvement of the outcome of pregnancy using fetal treatment.
Multimedia
| Media file 1: Prenatal diagnosis for congenital malformations and genetic disorders. Karyotype showing normal male chromosomal constitution (46, XY). |
| Media file 2: Prenatal diagnosis for congenital malformations and genetic disorders. Karyotype showing trisomy 21 (47, XY, +21) in a male. |
| Media file 3: Prenatal diagnosis for congenital malformations and genetic disorders. Segregation of haplotypes for 10 markers (M1-M10) in a family. Diseased haplotype, as indicated by red bars, is shared by all of the affected individuals (filled circles and squares) and by none of the unaffected individuals (unfilled circles and squares). |
 References
|